Reproducible analysis: package development
Issue
Packages are used to share your R code with others. Why then should this be the first topic tackled? Couldn’t we make our code shareable after we made something to actually share? Well, I actually don’t make the analysis a package mainly for sharing it (even though it’s nice to have). Packages also have the very nice side effect they enforce conventions. Other packages (e.g. testthat) build on these conventions. If we adhere to these conventions we’ll have way less issues interfacing with other people’s code. Conventions of course have an impact on nearly everything so it makes sense to make our analysis a package from the start.
Solution
In the R ecosystem devtools is the de facto standard to build packages. devtools helps with common tasks like scaffolding, running tests, setting up licenses, … I base myself mainly on the explanation in the R packages book. References to this book will popup later on as well when talking about tests or documentation.
At the moment there are 2 slightly different versions of the R packages book available. The first version is a bit outdated (published in April 2015) focusing on an older version of devtools. The second version is based on devtools 2.0.0. It’s more up to date but is still being written at the moment. I try as much as possible to work with the second edition. If some part is not yet available in the second edition I base myself on the first edition.
Creating package
To use devtools to create a package we first have to load it in the console:
library(devtools)
Let’s go to the root of our reproducible-analysis repository and run the following in the console to create a package:
create_package("reproseries")
This doesn’t work:

At first I tried to come up with ways to turn existing code into an R package. I was sorely disappointed by the lack of documentation on that subject. However, this is probably a good thing since it forces you to start from a clean slate which is what I did. Now the reproducible-analysis repository is abandoned in favor of reproseries.
Instead of running in the root of reproducible-analysis run this where you want to create the package:
create_package("reproseries")
Since there was no directory reproseries yet this works fine. Some files and directories are created automatically of which the most important are:
- R
- DESCRIPTION
- .gitignore
The R directory contains the functions used in the analysis, the DESCRIPTION file provides metadata about the package: its title, a short description of what it does, its version, …
Note the .gitignore file being there doesn’t mean the package is a Git repository yet.
Use Git
To make our package Git compatible, run use_git() in the console. Git and version control will be covered in a later post but what this command does is:
- initialize repo
- add all files to staging
- commit them as first commit
I could have done the same on the command line:
git initgit add -Agit commit -m "Initial commit"
However, why not use shortcuts if they’re available and clear?
Migrate functions to R
Last post we already wrote some functions for our analysis. It’s time to migrate these from the reproducible-analysis repository to the reproseries package.
Now is a good moment to think about how to structure these functions. Maybe we want to group them by the data analysis step they belong to (import-functions.R, tidy-functions.R, …)? Or maybe group them by the data they manipulate (temperatures.R, avg_temperatures.R)? It’s not a clear-cut choice but I noticed from experience consistency is key. Think about the grouping at the beginning of the project and stick with it not to confuse yourself. It’s very tempting to half way start rearranging everything. Only do if it’s really necesary and you know where you want to end up. In this case I think it makes a lot of sense to group by data analysis step.
The philosophy of devtools is to let helper functions do the heavy lifting for you. So instead of copying the code ourselves to the R folder let’s get some help from the use_r function. Running use_r("tidy") opens the tidy.R file in the R folder:
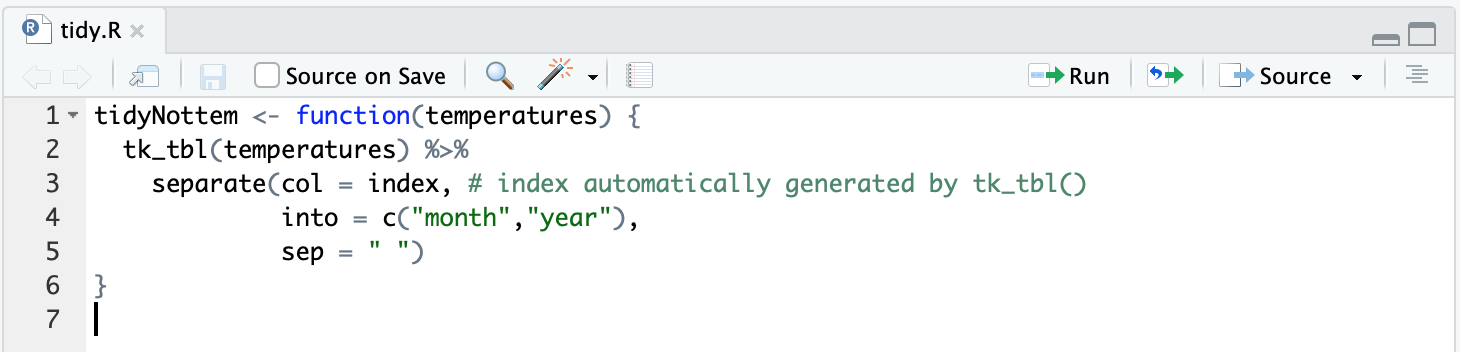
You might notice the tidyNottem function uses two other functions, tk_tbl from the timetk package and separate from the tidyr package (part of tidyverse). In the original analysis.R script we defined these dependencies with library. However, you don’t do it that way in a package. Packages are loaded in two distinct steps:
- build
- the actual loading
If you add a library call in your functions file this will only make the package available in the build phase but not in the actual load phase. To have the package available in the load phase you have to add it to the DESCRIPTION file with the Imports field:
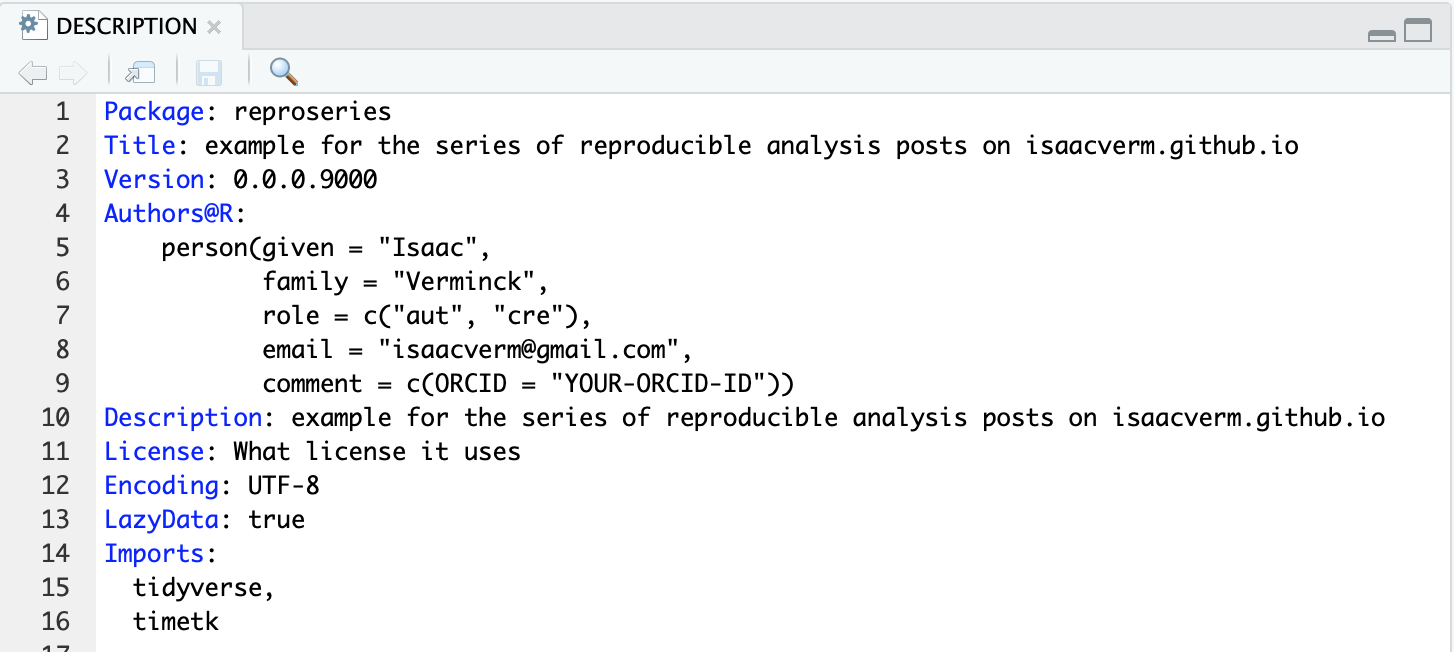
Note magrittr has to be imported to have access to the %>% pipe operator. Modify the NAMESPACE adding adding an importFrom call. I actually have no idea why it works with the NAMESPACE but didn’t by just adding it to imports in the DESCRIPTION. The NAMESPACE section of R packages clearly states:
NAMESPACE also controls which external functions can be used by your package without having to use ::.
Oh well.
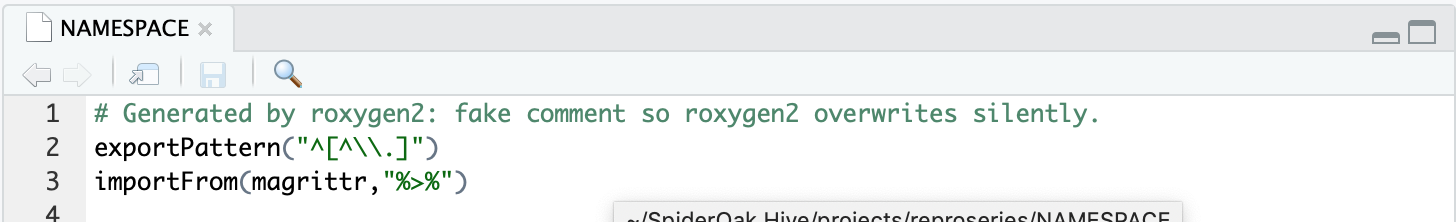
Other fields like license I leave untouched at the moment. They’re created by default but I don’t mess with them if not needed. You can see keeping tracks of external packages used can quickly become a hassle. At the moment I still remember separate belongs to the tidyverse and tk_tbl is part of timetk. I’m quite sure I won’t remember this in a few days, I dread talking about what I’ll remember about it in a few months. So it’s best practice to add the package name to the function name like this:
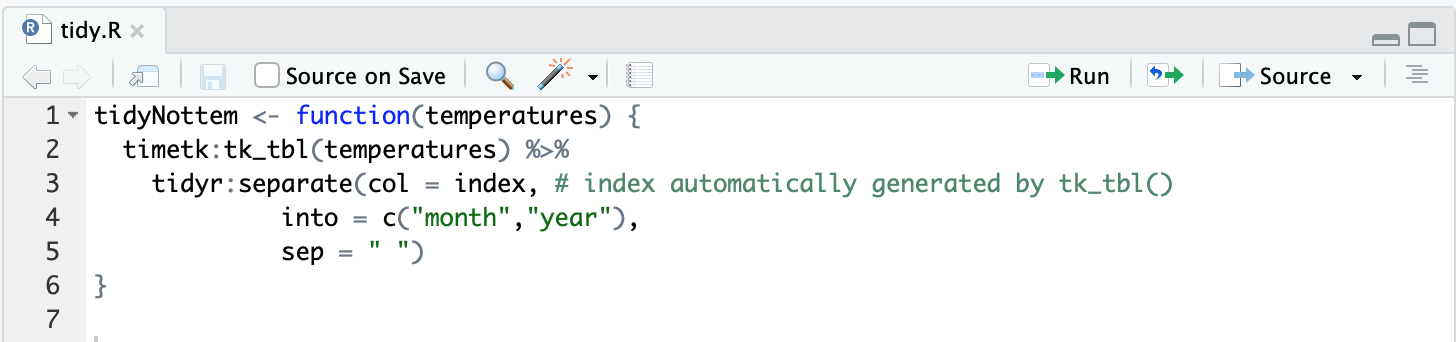
This does get verbose. An alternative solution would be to use namespace imports but that would lead us too far. In general I don’t mind it makes the function calls long. Try to look at it on the bright side. Since you lose a number of characters by default at the beginning you have to think harder about concise and practical function names.
Now let’s try if our tidy function does something by running this in the console:
library(devtools)
load_all()
tidyNottem(nottem)
load_all is a devtools function making all functions in the package available for interactive use. nottem is a built-in dataset so need to import it explicitly.
The process of migrating the other functions follows the same patterns as for the tidy function:
use_r()to create file- copy function definition to file (but no
librarycalls) load_allto test if function is available
If you want to check which functions are available after loading:
lsf.str("package:reproseries")
Check your build
Using check() you get some tests for free. check() will do some basic sanity checks. In my case it complained about the locale, description field and the license. All of these were easy to fix and I would have never found out about them myself. Actually things were even easier than I thought. I tried to find the solution for fixing the LICENSE field all by myself but there’s actually already a devtools function called use_mit_license() creating the exact setup we need.
There’s also an issue with using non-standard evaluation like the tidyverse packages do:
checking R code for possible problems ... NOTE
addCelsiusColumn: no visible binding for global variable ‘value’
calculateAutoCorrelation: no visible global function definition for
‘acf’
...
A solution is available but it has some disadvantages:
- you have to add
.datain pipes instead of using the implicit argument - it means rewriting all existing functions
I’d rather wait for an official fix in the tidyverse itself.
There’s one remaining issue namely the current lack of documentation. This will be handled in later posts:
❯ checking for missing documentation entries ... WARNING
Undocumented code objects:
‘addCelsiusColumn’ ‘calculateAutoCorrelation’
‘plotAvgTemperaturesByYear’ ‘summariseAvgTemperatureByYear’
‘tidyNottem’
All user-level objects in a package should have documentation entries.
In any case running check() regularly is a good sanity check!
Install your own package
Running load_all() just simulated loading the package. If you want the real thing run install(). Afterwards you can use your own package in any script by using library calls as before.
This is how our analysis script would look like:
library(reproseries)
# remove previous objects in global environment
rm(list = ls())
# objects
temperatures <- nottem %>%
tidyNottem %>%
...
As you can see:
- there are no
library(tidyverse), … calls anymore - the functions are in the package so no need to define them in the analysis
Summary
devtools provide handy helper functions for all the important parts of creating a package. Get started with create_package() to create a package. Do this from scratch and not an existing directory. Run use_git() to quickly get a repo set up. You can test interactively by running load_all() to simulate how your package will be loaded, run install() if you’re satisfied with the results and want to install your package locally. And remember to run check() as many times as possible to see if there are any issues to fix.
We now have a decent scaffolding to add some tests in the next post.